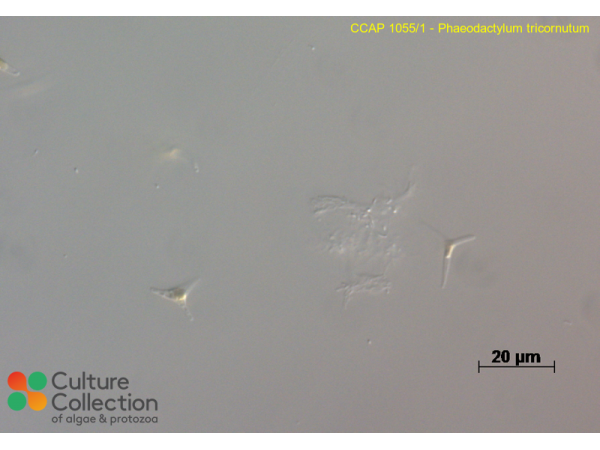
References [ 103 ]
Oudot-Le Secq MP, Grimwood J, Shapiro H, Armbrust EV, Bowler C & Green BR (2007) Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Molecular Genetics and Genomics 277: 427-439.
Bowler C et al. (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature (Letters) 456: -.
Gachon CMM, Day JG, Campbell CN, Pröschold T, Saxon RJ & Küpper FC (2007) The Culture Collection of Algae and Protozoa (CCAP): A biological resource for protistan genomics Gene 406: 51-57.
De Martino A, Meichenin A, Shi J, Pan K & Bowler C (2007) Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions. Journal of Phycology 43: 992-1009.
Huysman MJJ, Martens C, Vandepoele K, Gillard J, Rayko E, Heijde M, Bowler C, Inzé D, Van de Peer Y, De Veylder L & Vyverman W (2010) Genome-wide analysis of the diatom cell cycle unveils a novel type of cyclins involved in environmental signaling. Genome Biology 11: R17.
Montsant A, Maheswari U, Bowler C & Lopez PJ (2005) Diatomics: Toward diatom functional genomics. Journal of Nanoscience and Nanotechnology 5: 1-11.
Toda H & Itoh N (2011) Isolation and characterization of a gene encoding a S-adenosyl-L-methionine-dependent halide/thiol methyltransferase (HTMT) from the marine diatom Phaeodactylum tricornutum: Biogenic mechanism of CH3I emissions in oceans. Phytochemistry 72: 337-343.
De Martino A, Bartual A, Willis A, Meichenin A, Villazán B, Maheswari U & Bowler C (2011) Physiological and molecular evidence that environmental changes elicit morphological interconversion in the model diatom Phaeodactylum tricornutum. Protist 162: 462-481.
Yang EC, Boo GH, Kim HJ, Cho SM, Boo SM, Andersen RA & Yoon HS (2012) Supermatrix data highlight the phylogenetic relationships of photosynthetic stramenopiles. Protist 163: 217-231.
Shlykov MA, Zheng WH, Chen JS & Saier MH (2012) Bioinformatic characterization of the 4-Toluene Sulfonate Uptake Permease (TSUP) family of transmembrane proteins. Biochimica & Biophysica Acta 1818: 703-717.
Lei QY & Lu SH (2011) Molecular ecological responses of the dinoflagellate Karenia mikimotoi to phosphate stress. Harmful Algae 12: 39-45.
Momand J, Villegas A & Belyi VA (2011) The evolution of MDM2 family genes. Gene 486: 23-30.
Yu ET, Zendejas FJ, Lane PD, Gaucher S, Simmons BA & Lane TW (2009) Triacylglycerol accumulation and profiling in the model diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum (Baccilariophyceae) during starvation Journal of Applied Phycology 21: 669-681.
Lommer M, Specht M, Roy A, Kraemer L, Andreson R, Gutowska MA, Wolf J, Bergner SV, Schilhabel MB, Klostermeier UC, Beiko RG, Rosenstiel P, Hippler M & LaRoche J (2012) Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biology 13: R66.
Lopez Barreiro D, Zamalloa C, Boon N, Vyverman W, Ronsse F, Brilman W & Prins W (2013) Influence of strain-specific parameters on hydrothermal liquefaction of microalgae Bioresource Technology 146: 463-471.
Lavaud J & Lepetit B (2013) An explanation for the inter-species variability of the photoprotective non-photochemical chlorophyll fluorescence quenching in diatoms. Biochimica & Biophysica Acta 1827: 294-302.
Oncel SS (2013) Microalgae for a macroenergy world. Renewable and Sustainable Energy Reviews 26: 241-264.
Johnson EA & Lecomte JTJ (2013) Chapter Six - The globins of cyanobacteria and algae. Advances in Microbial Physiology 63: 195-272.
Vlachakis D, Pavlopoulou A, Kazazi D & Kossida S (2013) Unraveling microalgal molecular interactions using evolutionary and structural bioinformatics. Gene 528: 109-119.
Gangaplara A, Massilamany C, Steffen D & Reddy J (2013) Mimicry epitope from Ehrlichia canis for interphotoreceptor retinoid-binding protein 201-216 prevents autoimmune uveoretinitis by acting as altered peptide ligand. Journal of Neuroimmunology 263: 98-107.
Seligmann H (2013) Systematic asymmetric nucleotide exchanges produce human mitochondrial RNAs cryptically encoding for overlapping protein coding genes. Journal of Theoretical Biology 324: 1-20.
Lee MA, Faria DG, Han MS, Lee J & Ki JS (2013) Evaluation of nuclear ribosomal RNA and chloroplast gene markers for the DNA taxonomy of centric diatoms. Biochemical Systematics and Ecology 50: 163-174.
Perez-Lopez P, Gonzalez-Garcia S, Allewaert C, Verween A, Murray P, Feijoo G & Moreira MT (2014) Environmental evaluation of eicosapentaenoic acid production by Phaeodactylum tricornutum. Science of the Total Environment 466-467: 991-1002.
Formighieri C, Cazzaniga S, Kuras R & Bassi R (2013) Biogenesis of photosynthetic complexes in the chloroplast of Chlamydomonas reinhardtii requires ARSA1, a homolog of prokaryotic arsenite transporter and eukaryotic TRC40 for guided entry of tail-anchored proteins. The Plant Journal 73: 850-861.
Zhuang Y, Zhang H & Lin S (2013) Polyadenylation of 18S rRNA in algae Journal of Phycology 49: 570-579.
Rahman MM, Rahman MA, Maki T, Nishiuchi T, Asano T & Hasegawa H (2014) A marine phytoplankton (Prymnesium parvum) up-regulates ABC transporters and several other proteins to acclimatize with Fe-limitation. Chemosphere 95: 213-219.
Zhang C, Lin S, Huang L, Lu W, Li M & Liu S (2014) Suppression subtraction hybridization analysis revealed regulation of some cell cycle and toxin genes in Alexandrium catenella by phosphate limitation. Harmful Algae 39: 26-39.
Fabris M, Matthijs M, Carbonelle S, Moses T, Pollier J, Dasseville R, Baart GJE, Wyverman W & Goossens A (2014) Tracking the sterol biosynthesis pathway of the diatom Phaeodactylum tricornutum. New Phytologist 204: 521-535.
Cuvelier ML, Allen AE, Monier A, McCrow JP, Messie M, Tringe SG, Woyke T, Welsh RM, Ishoey T, Lee JH, Binder BJ, DuPont CL, Latasa M, Guigand C, Buck KR, Hilton J, Thiagarajan M, Caler E, Read B, Lasken RS, Chavez FP & Worden AZ (2010) Targeted metagenomics and ecology of globally important uncultured eukaryotic phytoplankton. PNAS 107: 14679-14684.
He L, Han X & Yu Z (2014) A rare Phaeodactylum tricornutum cruciform morphotype: Culture conditions, transformation and unique fatty acid characteristics. PLoS ONE 9: e93922.
Nakayama T, Ishida KI & Archibald JM (2012) Broad distribution of TPI-GAPDH fusion proteins among eukaryotes: Evidence for glycolytic reactions in the mitochondrion? PLoS ONE 7(12): e52340.
Qian D, Jiang L, Lu L, Wei C & Li Y (2011) Biochemical and structural properties of cyanases from Arabidopsis thaliana and Oryza sativa. PLoS ONE 6(3): e18300.
Tsaousis AD, Gentekaki E, Eme L, Gaston D & Roger AJ (2014) Evolution of the cytosolic iron-sulfur cluster assembly machinery in Blastocystis species and other microbial eukaryotes. Eukaryotic Cell 13: 143-153.
Karkar S, Facchinelli F, Price DC, Weber APM & Bhattacharya (2015) Metabolic connectivity as a driver of host and endosymbiont integration. PNAS 112 (33): 10208-10215.
Maruyama S, Shoguchi E, Satoh N & Minagawa J (2015) Diversification of the light-harvesting complex gene family via intra- and intergenic duplications in the coral symbiotic alga Symbiodinium. PLoS ONE 10: e0119406.
Schulze B, Buhmann MT, Río Bártulos C & Kroth PG (2015) Comprehensive computational analysis of leucine-rich repeat (LRR) proteins encoded in the genome of the diatom Phaeodactylum tricornutum. Marine Genomics 21: 43-51.
Xiang T, Nelson W, Rodriguez J, Tolleter D & Grossman AR (2015) Symbiodinium transcriptome and global responses of cells to immediate changes in light intensity when grown under autotrophic or mixotrophic conditions. The Plant Journal 82: 67-80.
Stanley MS & Callow JA (2007) Whole cell adhesion strength of morphotypes and isolates of Phaeodactylum tricornutum (Bacillariophyceae). European Journal of Phycology 42: 191-197.
Zhu BH, Shi HP, Yang GP, Lv NN, Yang M & Pan KH (2015) Silencing UDP-glucose pyrophosphorylase gene in Phaeodactylum tricornutum affects carbon allocation. New Biotechnology 33: 237-244.
Réveillon D, Séchet V, Hess P & Amzil Z (2015) Systematic detection of BMAA (β-N-methylamino-L-alanine) and DAB (2,4-diaminobutyric acid) in mollusks collected in shellfish production areas along the French coasts. Toxicon 110: 35-46.
Réveillon D, Abadie E, Séchet v, Masseret E, Hess P & Amzil Z (2015) β-N-methylamino-L-alanine (BMAA) and isomers: Distribution in different food web compartments of Thau lagoon, French Mediterranean Sea. Marine Environmental Research 110: 8-18.
Laohavisit A, Anderson A, Bombelli P, Jacobs M, Howe CJ, Davies JM & Smith AG (2015) Enhancing plasma membrane NADPH oxidase activity increases current output by diatoms in biophotovoltaic devices Algal Research 12: 91-98.
Bailleul B, Berne N, Murik O, Petroutsos D, Prihoda J, Tanaka A, Villanova V, Bligny R, Flori S, Falconet D, Krieger-Liszkay A, Santabarbara S, Rappaport F, Joliot P, Tirichine L, Falkowski PG, Cardol P, Bowler C & Finazzi G (2015) Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524: 366.
Mabbitt PD, Wilbanks SM & Eaton-Rye JJ (2014) Duplication and divergence of the Psb27 subunit of Photosystem II in the green algal lineage. New Zealand Journal of Botany 52: 74-83.
Buhmann MT, Schulze B, Förderer A, Schleheck D & Kroth PG (2016) Bacteria may induce the secretion of mucin-like proteins by the diatom Phaeodactylum tricornutum. Journal of Phycology 52: 463-474.
Longworth J, Wu D, Huete-Ortega M, Wright PC & Vaidyanathan S (2016) Proteome response of Phaeodactylum tricornutum, during lipid accumulation induced by nitrogen depletion. Algal Research 18: 213-224.
Lama S, Muylaert K, Karki TB, Foubert I, Henderson RK & Vandamme D (2016) Flocculation properties of several microalgae and a cyanobacterium species during ferric chloride, chitosan and alkaline flocculation. Bioresource Technology 220: 464-470.
Shen Q, Chen Y, Jun D, Lin H, Wang Q & Zhao YH (2016) Comparative genome analysis of the oleaginous yeast Trichosporon fermentans reveals its potential application in lipid accumulation. Microbiological Research 192: 203-210.
Jallet D, Caballero MA, Gallin AA, Youngblood M & Peers G (2016) Photosynthetic physiology and biomass partitioning in the model diatom Phaeodactylum tricornutum grown in a sinusoidal light regime. Algal Research 18: 51-60.
Caballero MA, Jallet D, Shi L, Rithner C, Zhang Y & Peers G (2016) Quantification of chrysolaminarin from the model diatom Phaeodactylum tricornutum. Algal Research 20: 180-188.
Réveillon D, Séchet V, Hess P & Amzil Z (2016) Production of BMAA and DAB by diatoms (Phaeodactylum tricornutum, Chaetoceros sp., Chaetoceros calcitrans and, Thalassiosira pseudonana) and bacteria isolated from a diatom culture. Harmful Algae 58: 45-50.
Wang H, Mi T, Zhen Y, Jing X, Liu Q & Yu Z (2017) Metacaspases and programmed cell death in Skeletonema marinoi in response to silicate limitation. Journal of Plankton Research -: 1-15.
Andrés-León E, Cases I, Arcas A & Rojas AM (2016) DDRprot: a database of DNA damage response-related proteins. Database -: 1-10.
Lepetit B, Gélin G, Lepetit M, Sturm S, Vugrinec S, Rogato A, Kroth PG, Falciatore A & Lavaud J (2017) The diatom Phaeodactylum tricornutum adjusts nonphotochemical fluorescence quenching capacity in response to dynamic light via fine-tuned Lhcx and xanthophyll cycle pigment synthesis New Phytologist 214: 205-218.
Serif M, Lepetit B, Weißert K, Kroth PG & Rio Bartulos C (2017) A fast and reliable strategy to generate TALEN-mediated gene knockouts in the diatom Phaeodactylum tricornutum. Algal Research 23: 186-195.
Arakaki A, Matsumoto T, Tateishi T, Matsumoto M, Nojima D, Tomoko Y & Tanaka T (2017) UV-C irradiation accelerates neutral lipid synthesis in the marine oleaginous diatom Fistulifera solaris Bioresource Technology 245: 1520-1526.
Vandamme D, Gheysen L, Muylaert K & Foubert I (2018) Impact of harvesting method on total lipid content and extraction efficiency for Phaeodactylum tricornutum Separation and Purification Technology 194: 362-367.
Gutowska MA, Shome B, Sudek S, McRose DL, Hamilton M, Giovannoni SJ, Begley TP & Worden AZ (2017) Globally Important Haptophyte Algae Use Exogenous Pyrimidine Compounds More Efficiently than Thiamin mBio 8: e01459-17.
Du N, Gholami P, Kline DI, DuPont CL, Dickson AG, Mendola D, Martz T, Allen AE & Mitchell BG (2018) Simultaneous quantum yield measurements of carbon uptake and oxygen evolution in microalgal cultures PLoS ONE 13: e0199125.
Han KY, Graf L, Reyes CP, Melkonian B, Andersen RA, Yoon HS & Melkonian M (2018) A Re-investigation of Sarcinochrysis marina (Sarcinochrysidales, Pelagophyceae) from its Type Locality and the Descriptions of Arachnochrysis, Pelagospilus, Sargassococcus and Sungminbooa genera nov. Protist 169: 79-106.
Singh A, Tyagi C, Nath O & Singh IK (2018) Helicoverpa-inducible Thioredoxin h from Cicer arietinum: structural modeling and potential targets International Journal of Biological Macromolecules 109: 231-243.
Mix AK, Cenci U, Heimerl T, Marter P, Wirkner ML & Moog D (2018) Identification and Localization of Peroxisomal Biogenesis Proteins Indicates the Presence of Peroxisomes in the Cryptophyte Guillardia theta and Other "Chromalveolates" Genome Biology and Evolution 10: 2834-2852.
Havurinne V &Tyystjärvi E (2017) Action Spectrum of Photoinhibition in the Diatom Phaeodactylum tricornutum Plant & Cell Physiology 58: 2217-2225.
König S, Eisenhut M, Bräutigam A, Kurz S, Weber APM & Büchel C (2017) The Influence of a Cryptochrome on the Gene Expression Profile in the Diatom Phaeodactylum tricornutum under Blue Light and in Darkness Plant & Cell Physiology 58: 1914-1923.
Yang M, Lin X, Liu X, Zhang J & Ge F (2018) Genome Annotation of a Model Diatom Phaeodactylum tricornutum Using an Integrated Proteogenomic Pipeline Molecular Plant 11: 1292-1307.
Broddrick JT, Du N, Smith SR, Tsuji Y, Jallet D, Ware MA, Peers G, Matsuda Y, Dupont CL, Mitchell BG, Palsson BO & Allen AE (2019) Cross-compartment metabolic coupling enables flexible photoprotective mechanisms in the diatom Phaeodactylum tricornutum New Phytologist -: -.
Bernal-Bayard P, Alvarez C, Calvo P, Castell C, Roncel M, Hervás M & Navarro JA (2019) The singular properties of photosynthetic cytochrome c550 from the diatom Phaeodactylum tricornutum suggest new alternative functions Physiologia Plantarum -: -.
Bojko M, Olchawa-Pajor M, Goss R, Schaller-Laudel S, Strzalka K & Latowski D (2018) Diadinoxanthin de‐epoxidation as important factor in the short‐term stabilization of diatom photosynthetic membranes exposed to different temperatures Plant, Cell and Environment -: -.
Havurinne V, Mattila H, Antinluoma M & Tyystjärvi E (2018) Unresolved quenching mechanisms of chlorophyll fluorescence may invalidate MT saturating pulse analyses of photosynthetic electron transfer in microalgae Physiologia Plantarum -: -.
Lupette J, Jaussaud A, Seddiki K, Marabito C, Brugière S, Schaller H, Kuntz M, Putaux JL, Jouneau PH, Rébeillé F, Falconet D, CoutéY, Jouhet J, Tardif M, Salvaing J & Maréchal E (2019) The architecture of lipid droplets in the diatom Phaeodactylum tricornutum Algal Research 38: 101415.
Sun L, Chin WC, Chiu MH, Xu C, Lin P, Schwehr KA, Quigg A & Santschi PH (2019) Sunlight induced aggregation of dissolved organic matter: Role of proteins in linking organic carbon and nitrogen cycling in seawater Science of the Total Environment 654: 872-877.
Santaeufemia S, Abalde J & Torres E (2019) Eco-friendly rapid removal of triclosan from seawater using biomass of a microalgal species: Kinetic and equilibrium studies Journal of Hazardous Materials -: -.
Bowler C & Falciatore A (2019) Genome of the Month: Phaeodactylum tricornutum Trends in Genetics 35: 706-707.
Smith SR, Dupont CL, McCarthy JK, Broddrick JT, Oborník M, Horák A, Füssy Z, Cihlár J, Kleessen S, Zheng H, McCrow JP, Hixson KK, Araújo WL, Nunes-Nesi A, Fernie A, Nikoloski Z, Palsson BO & Allen AE (2019) Evolution and regulation of nitrogen flux through compartmentalized metabolic networks in a marine diatom Nature Communications 10: 4552.
Nef C, Jung S, Mairet F, Kaas R, Grizeau D & Garnier M (2019) How haptophytes microalgae mitigate vitamin B12 limitation Scientific Reports 9: 8417.
Buck JM, Sherman J, Río Bártulos C, Serif M, Halder M, Henkel J, Falciatore A, Lavaud J, Gorbunov M, Kroth PG, Falkowski PG & Lepetit B (2019) Lhcx proteins provide photoprotection via thermal dissipation of absorbed light in the diatom Phaeodactylum tricornutum Nature Communications 10: 4167.
Brumwell SL, MacLeod MR, Huang T, Cochrane RR, Meaney RS, Zamani M, Matysiakiewicz O, Dan KN, Janakirama P, Edgell DR, Charles TC, Finan TM & Karas BJ (2019) Designer Sinorhizobium meliloti strains and multi-functional vectors enable direct inter-kingdom DNA transfer PLoS ONE 14: e0206781.
Anto S, Karpagam R, Renukadevi P, Jawaharraj K & Varalakshmi P (2019) Biomass enhancement and bioconversion of brown marine microalgal lipid using heterogeneous catalysts mediated transesterification from biowaste derived biochar and bionanoparticle Fuel 255: 115789.
Thakur R, Shiratori T & Ishida K (2019) Taxon-rich Multigene Phylogenetic Analyses Resolve the Phylogenetic Relationship Among Deep-branching Stramenopiles Protist 170: 125682.
Ganuza E, Yang S, Amezquita M, Giraldo-Silva A & Andersen RA (2019) Genomics, Biology and Phylogeny Aurantiochytrium acetophilum sp. nov. (Thraustrochytriaceae), Including First Evidence of Sexual Reproduction Protist 170: 209-232.
Sugiyama K, Takahashi K, Nakazawa K, Yamada M, Kato S, Shinomura T, Nagashima Y, Suzuki H, Ara T, Harada J & Takaichi S (2019) Oxygenic Phototrophs Need ζ-Carotene Isomerase (Z-ISO) for Carotene Synthesis: Functional Analysis in Arthrospira and Euglena Plant & Cell Physiology 61: 276-282.
Butler T, Kapoore RV & Vaidyanathan S (2020) Phaeodactylum tricornutum: A diatom cell factory Trends in Biotechnology -: -.
Morabito C, Bournaud C, Maës C, Schuler M, Aiese Cigliano R, Dellero Y, Maréechal E, Amato A & Rébeillé (2019) The lipid metabolism in thraustochytrids Progress in Lipid Research 76: 101007.
González-Aravena M, Kenny NJ, Osorio M, Font A, Riesgo A & Cárdenas CA (2019) Warm temperatures, cool sponges: the effect of increased temperatures on the Antarctic sponge Isodictya sp. PeerJ 7: e8088.
Penhaul Smith JK, Beveridge C, Laudicella VA, Hughes AD, McEvoy L & Day JG (2021) Utilising mixotrophically cultured "designer algae" as blue mussel larval feed. Aquaculture International -: -.
Butler TO, Acurio K, Mukherjee J, Dangasuk MM, Corona O & Vaidyanathan S (2021) The transition away from chemical flocculants: Commercially viable harvesting of Phaeodactylum tricornutum Separation and Purification Technology 255: 117733.
Song Z, Lye GJ & Parker BM (2020) Morphological and biochemical changes in Phaeodactylum tricornutum triggered by culture media: Implications for industrial exploitation Algal Research 47: 101822.
Yoshino T, Mao Y, Maeda Y, Negishi R, Murata S, Moriya S, Shimada H, Arakaki A, Kobayashi K, Hagiwara Y, Okamoto K & Tanaka T (2022) Single-cell genotyping of phytoplankton from ocean water by gel-based cell manipulation Biotechnology Journal 17(6): 2100633.
Pandey A, Kant G, Afzal S, Singh MP, Singh NK, Kumar S & Srivastava S (2022) Chapter 5 - Genetic manipulation of microalgae for enhanced biotechnological applications In: Handbook of Algal Biofuels -: 97-122.
Yu Z, Geisler K, Leontidou T, Young REB, Vonlanthen SE, Purton S, Abell C & Smith AG (2021) Droplet-based microfluidic screening and sorting of microalgal populations for strain engineering applications Algal Research 56: 102293.
Penhaul Smith JK, Hughes AD, McEvoy L & Day JG (2023) Use of crude glycerol for mixotrophic culture of Phaeodactylum tricornutum. Algal Research 69: 102929.
Rumin J, Carrier G, Rouxel C, Charrier A, Raimbault V, Cadoret JP, Bougaran G & Saint-Jean B (2023) Towards the optimization of genetic polymorphism with EMS-induced mutagenesis in Phaeodactylum tricornutum Algal Research 74: 103148.
Su Y, Xu M, Brynjólfsson S & Fu W (2023) Physiological and molecular insights into adaptive evolution of the marine model diatom Phaeodactylum tricornutum under low-pH stress Journal of Cleaner Production 412: 137297.
Sayer AP, Llavero-Pasquina M, Geisler K, Holzer A, Bunbury F, Mendoza-Ochoa GI, Lawrence AD, Warren MJ, Mehrshahi P & Smith AG (2023) Conserved cobalamin acquisition protein 1 is essential for vitamin B12 uptake in both Chlamydomonas and Phaeodactylum. Plant Physiology kiad564: -.
Reynolds-Brandao P, Quintas-Nunes F, Bertrand CDF, Martins RM, Crespo MTB, Galinha CF & Nascimento FX (2025) Integration of spectroscopic techniques and machine learning for optimizing Phaeodactylum tricornutum cell and fucoxanthin productivity. Bioresource Technology 418: 131988.
Yun HS, Yoneda K, Sugasawa T, Suzuki I & Maeda Y (2024) Genome-wide mapping of autonomously replicating sequences in the marine diatom Phaeodactylum tricornutum Marine Biotechnology -: -.
Bertrand CDF, Martins R, Quintas-Nunes F, Reynolds-Brandão P, Barreto Crespo MT & Nascimento FX (2024) Exploring the functional and genomic features of Cellulophaga lytica NFXS1, a zeaxanthin and lytic enzyme-producing marine bacterium that promotes microalgae growth The Microbe 4: 100142.
Su Y, Chen J, Hu J, Qian C, Ma J, Brynjolfsson S & Fu W (2024) Manipulation of ion/electron carrier genes in the model diatom Phaeodactylum tricornutum enables its growth under lethal acidic stress iScience 27(8): 110482.
He Z, Wang J & Li Y (2024) Recent Advances in Microalgae-driven Carbon Capture, Utilization, and Storage: Strain Engineering through Adaptive Laboratory Evolution and Microbiome Optimization Green Carbon -: -.
Fantino E, Awwad F, Merindol N, Garza AMD, Gélinas SE, Robles GCG, Custeau A, Meddeb-Mouelhi F & Desgagné-Penix I (2024) Bioengineering Phaeodactylum tricornutum, a marine diatom, for cannabinoid biosynthesis Algal Research 77: 103379.
Zhao DS, Farooq MA, Li M, Chen YT, Xu JM, Liu XL, Zhang A, Yan X, Zou HX & Pang Q (2024) Acute toxicity of salicylic acid and its derivatives on the diatom Phaeodactylum tricornutum: Physico-Biochemical and transcriptomic insights Aquatic Toxicology 276: 107116.
Grujcic V, Saarenpää S, Sundh J, Sennblad B, Norgren B, Latz M, Giacomello S, Foster RA & Andersson AF (2024) Towards high-throughput parallel imaging and single-cell transcriptomics of microbial eukaryotic plankton PLoS ONE 19(1): e0296672.
Marchese D, Rosengart A, Barbera E & Sforza E (2025) Exploring a bicarbonate-based carbon capture approach applicability to industrially relevant marine microalgae. Chemical Engineering Transactions 117: 259-264.