(Bold text = submission by CCAP staff or collaborators)
Note: for strains where we have DNA barcodes we can be reasonably confident of identity, however for those not yet sequenced we rely on morphology
and the original identification, usually made by the depositor. Although CCAP makes every effort to ensure the correct taxonomic identity of strains, we cannot guarantee
that a strain is correctly identified at the species, genus or class levels. On this basis users are responsible for confirming the identity of the strain(s) they receive
from us on arrival before starting experiments.
For strain taxonomy we generally use AlgaeBase for algae and
Adl et al. (2019) for protists.
| Attributes | |
| Authority | Dangeard 1888 |
| Isolator | Smith |
| Collection Site | potato field Amherst, Massachusetts, USA |
| Notes | this strain carries the nit1 and nit2 mutations and cannot grow on nitrate as the sole N source (via www.chlamycollection.org record for 11/32C). |
| Axenicity Status | Axenic |
| Area | North America |
| Country | USA |
| Environment | Soil |
| GMO | No |
| In Scope of Nagoya Protocol | No |
| ABS Note | Collected pre Nagoya Protocol. No known Nagoya Protocol restrictions for this strain. |
| Collection Date | pre. 1961 |
| Original Designation | 137C+ |
| Pathogen | Not pathogenic: Hazard Class 1 |
| Strain Maintenance Sheet | SM_GeneralFreshwaterGreens.pdf |
| Toxin Producer | Not Toxic / No Data |
| Type Culture | No |
| Taxonomy WoRMS ID | 608420 |
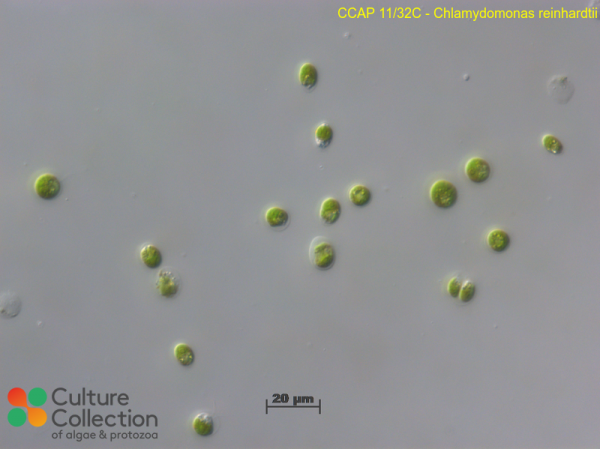
CCAP 11/32C
Chlamydomonas reinhardtii
- Product Code: CCAP 11/32C
- Availability: See Availability/Lead Times
Related Products
CCAP FAEGJM-C
EG:JM Medium
CONCENTRATED STOCKS
Non-sterile concentrated stocks to make up 5 litres of EG:JM medium (a 1:1 mix of EG and JM media).
CCAP FAEGJM-P
EG:JM Medium
1 LITRE PREMADE
1 litre of sterile, ready to use, EG:JM medium. EG:JM medium is used for culturing freshwater algae